update from Github version of CoastSat
parent
b32d7d1239
commit
417fe12923
@ -0,0 +1,31 @@
|
||||
---
|
||||
name: Bug report
|
||||
about: Create a report to help us improve
|
||||
title: ''
|
||||
labels: bug
|
||||
assignees: ''
|
||||
|
||||
---
|
||||
|
||||
**Describe the bug**
|
||||
A clear and concise description of what the bug is.
|
||||
|
||||
**To Reproduce**
|
||||
Steps to reproduce the behavior (you can also attach your script):
|
||||
1. Go to '...'
|
||||
2. Click on '....'
|
||||
3. Scroll down to '....'
|
||||
4. See error
|
||||
|
||||
**Expected behavior**
|
||||
A clear and concise description of what you expected to happen.
|
||||
|
||||
**Screenshots**
|
||||
If applicable, add screenshots to help explain your problem.
|
||||
|
||||
**Desktop (please complete the following information):**
|
||||
- OS: [e.g. iOS]
|
||||
- CoastSat Version [e.g. 22]
|
||||
|
||||
**Additional context**
|
||||
Add any other context about the problem here.
|
||||
@ -0,0 +1,20 @@
|
||||
---
|
||||
name: Feature request
|
||||
about: Suggest an idea for this project
|
||||
title: ''
|
||||
labels: enhancement
|
||||
assignees: ''
|
||||
|
||||
---
|
||||
|
||||
**Is your feature request related to a problem? Please describe.**
|
||||
A clear and concise description of what the problem is. Ex. I'm always frustrated when [...]
|
||||
|
||||
**Describe the solution you'd like**
|
||||
A clear and concise description of what you want to happen.
|
||||
|
||||
**Describe alternatives you've considered**
|
||||
A clear and concise description of any alternative solutions or features you've considered.
|
||||
|
||||
**Additional context**
|
||||
Add any other context or screenshots about the feature request here.
|
||||
Binary file not shown.
Binary file not shown.
Binary file not shown.
@ -0,0 +1,436 @@
|
||||
{
|
||||
"cells": [
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"# Train a new classifier for CoastSat\n",
|
||||
"\n",
|
||||
"In this notebook the CoastSat classifier is trained using satellite images from new sites. This can improve the accuracy of the shoreline detection if the users are experiencing issues with the default classifier."
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"#### Initial settings"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": [],
|
||||
"run_control": {
|
||||
"marked": false
|
||||
}
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# load modules\n",
|
||||
"%load_ext autoreload\n",
|
||||
"%autoreload 2\n",
|
||||
"import os, sys\n",
|
||||
"import numpy as np\n",
|
||||
"import pickle\n",
|
||||
"import warnings\n",
|
||||
"warnings.filterwarnings(\"ignore\")\n",
|
||||
"import matplotlib.pyplot as plt\n",
|
||||
"\n",
|
||||
"# sklearn modules\n",
|
||||
"from sklearn.model_selection import train_test_split\n",
|
||||
"from sklearn.neural_network import MLPClassifier\n",
|
||||
"from sklearn.model_selection import cross_val_score\n",
|
||||
"from sklearn.externals import joblib\n",
|
||||
"\n",
|
||||
"# coastsat modules\n",
|
||||
"sys.path.insert(0, os.pardir)\n",
|
||||
"from coastsat import SDS_download, SDS_preprocess, SDS_shoreline, SDS_tools, SDS_classify\n",
|
||||
"\n",
|
||||
"# plotting params\n",
|
||||
"plt.rcParams['font.size'] = 14\n",
|
||||
"plt.rcParams['xtick.labelsize'] = 12\n",
|
||||
"plt.rcParams['ytick.labelsize'] = 12\n",
|
||||
"plt.rcParams['axes.titlesize'] = 12\n",
|
||||
"plt.rcParams['axes.labelsize'] = 12\n",
|
||||
"\n",
|
||||
"# filepaths \n",
|
||||
"filepath_images = os.path.join(os.getcwd(), 'data')\n",
|
||||
"filepath_train = os.path.join(os.getcwd(), 'training_data')\n",
|
||||
"filepath_models = os.path.join(os.getcwd(), 'models')\n",
|
||||
"\n",
|
||||
"# settings\n",
|
||||
"settings ={'filepath_train':filepath_train, # folder where the labelled images will be stored\n",
|
||||
" 'cloud_thresh':0.9, # percentage of cloudy pixels accepted on the image\n",
|
||||
" 'cloud_mask_issue':True, # set to True if problems with the default cloud mask \n",
|
||||
" 'inputs':{'filepath':filepath_images}, # folder where the images are stored\n",
|
||||
" 'labels':{'sand':1,'white-water':2,'water':3,'other land features':4}, # labels for the classifier\n",
|
||||
" 'colors':{'sand':[1, 0.65, 0],'white-water':[1,0,1],'water':[0.1,0.1,0.7],'other land features':[0.8,0.8,0.1]},\n",
|
||||
" 'tolerance':0.01, # this is the pixel intensity tolerance, when using flood fill for sandy pixels\n",
|
||||
" # set to 0 to select one pixel at a time\n",
|
||||
" }\n",
|
||||
" \n",
|
||||
"# read kml files for the training sites\n",
|
||||
"filepath_sites = os.path.join(os.getcwd(), 'training_sites')\n",
|
||||
"train_sites = os.listdir(filepath_sites)\n",
|
||||
"print('Sites for training:\\n%s\\n'%train_sites)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"### 1. Download images\n",
|
||||
"\n",
|
||||
"For each site on which you want to train the classifier, save a .kml file with the region of interest (5 vertices clockwise, first and last points are the same, can be created from Google myMaps) in the folder *\\training_sites*.\n",
|
||||
"\n",
|
||||
"You only need a few images (~10) to train the classifier."
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": []
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# dowload images at the sites\n",
|
||||
"dates = ['2019-01-01', '2019-07-01']\n",
|
||||
"sat_list = 'L8'\n",
|
||||
"for site in train_sites:\n",
|
||||
" polygon = SDS_tools.polygon_from_kml(os.path.join(filepath_sites,site))\n",
|
||||
" sitename = site[:site.find('.')] \n",
|
||||
" inputs = {'polygon':polygon, 'dates':dates, 'sat_list':sat_list,\n",
|
||||
" 'sitename':sitename, 'filepath':filepath_images}\n",
|
||||
" print(sitename)\n",
|
||||
" metadata = SDS_download.retrieve_images(inputs)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"### 2. Label images\n",
|
||||
"\n",
|
||||
"Label the images into 4 classes: sand, white-water, water and other land features.\n",
|
||||
"\n",
|
||||
"The labelled images are saved in the *filepath_train* and can be visualised afterwards for quality control. If yo make a mistake, don't worry, this can be fixed later by deleting the labelled image."
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": [],
|
||||
"run_control": {
|
||||
"marked": true
|
||||
}
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# label the images with an interactive annotator\n",
|
||||
"%matplotlib qt\n",
|
||||
"for site in train_sites:\n",
|
||||
" settings['inputs']['sitename'] = site[:site.find('.')] \n",
|
||||
" # load metadata\n",
|
||||
" metadata = SDS_download.get_metadata(settings['inputs'])\n",
|
||||
" # label images\n",
|
||||
" SDS_classify.label_images(metadata,settings)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"### 3. Train Classifier\n",
|
||||
"\n",
|
||||
"A Multilayer Perceptron is trained with *scikit-learn*. To train the classifier, the training data needs to be loaded.\n",
|
||||
"\n",
|
||||
"You can use the data that was labelled here and/or the original CoastSat training data."
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# load labelled images\n",
|
||||
"features = SDS_classify.load_labels(train_sites, settings)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# you can also load the original CoastSat training data (and optionally merge it with your labelled data)\n",
|
||||
"with open(os.path.join(settings['filepath_train'], 'CoastSat_training_set_L8.pkl'), 'rb') as f:\n",
|
||||
" features_original = pickle.load(f)\n",
|
||||
"for key in features_original.keys():\n",
|
||||
" print('%s : %d pixels'%(key,len(features_original[key])))"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"Run this section to combine the original training data with your labelled data:"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": []
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# add the white-water data from the original training data\n",
|
||||
"features['white-water'] = np.append(features['white-water'], features_original['white-water'], axis=0)\n",
|
||||
"# or merge all the classes\n",
|
||||
"# for key in features.keys():\n",
|
||||
"# features[key] = np.append(features[key], features_original[key], axis=0)\n",
|
||||
"# features = features_original \n",
|
||||
"for key in features.keys():\n",
|
||||
" print('%s : %d pixels'%(key,len(features[key])))"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"[OPTIONAL] As the classes do not have the same number of pixels, it is good practice to subsample the very large classes (in this case 'water' and 'other land features'):"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# subsample randomly the land and water classes\n",
|
||||
"# as the most important class is 'sand', the number of samples should be close to the number of sand pixels\n",
|
||||
"n_samples = 5000\n",
|
||||
"for key in ['water', 'other land features']:\n",
|
||||
" features[key] = features[key][np.random.choice(features[key].shape[0], n_samples, replace=False),:]\n",
|
||||
"# print classes again\n",
|
||||
"for key in features.keys():\n",
|
||||
" print('%s : %d pixels'%(key,len(features[key])))"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"When the labelled data is ready, format it into X, a matrix of features, and y, a vector of labels:"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": [],
|
||||
"run_control": {
|
||||
"marked": true
|
||||
}
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# format into X (features) and y (labels) \n",
|
||||
"classes = ['sand','white-water','water','other land features']\n",
|
||||
"labels = [1,2,3,0]\n",
|
||||
"X,y = SDS_classify.format_training_data(features, classes, labels)"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"Divide the dataset into train and test: train on 70% of the data and evaluate on the other 30%:"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": [],
|
||||
"run_control": {
|
||||
"marked": true
|
||||
}
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# divide in train and test and evaluate the classifier\n",
|
||||
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, shuffle=True, random_state=0)\n",
|
||||
"classifier = MLPClassifier(hidden_layer_sizes=(100,50), solver='adam')\n",
|
||||
"classifier.fit(X_train,y_train)\n",
|
||||
"print('Accuracy: %0.4f' % classifier.score(X_test,y_test))"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"[OPTIONAL] A more robust evaluation is 10-fold cross-validation (may take a few minutes to run):"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": [],
|
||||
"run_control": {
|
||||
"marked": true
|
||||
}
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# cross-validation\n",
|
||||
"scores = cross_val_score(classifier, X, y, cv=10)\n",
|
||||
"print('Accuracy: %0.4f (+/- %0.4f)' % (scores.mean(), scores.std() * 2))"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"Plot a confusion matrix:"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {
|
||||
"code_folding": []
|
||||
},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# plot confusion matrix\n",
|
||||
"%matplotlib inline\n",
|
||||
"y_pred = classifier.predict(X_test)\n",
|
||||
"SDS_classify.plot_confusion_matrix(y_test, y_pred,\n",
|
||||
" classes=['other land features','sand','white-water','water'],\n",
|
||||
" normalize=False);"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"When satisfied with the accuracy and confusion matrix, train the model using ALL the training data and save it:"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# train with all the data and save the final classifier\n",
|
||||
"classifier = MLPClassifier(hidden_layer_sizes=(100,50), solver='adam')\n",
|
||||
"classifier.fit(X,y)\n",
|
||||
"joblib.dump(classifier, os.path.join(filepath_models, 'NN_4classes_Landsat_test.pkl'))"
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "markdown",
|
||||
"metadata": {},
|
||||
"source": [
|
||||
"### 4. Evaluate the classifier\n",
|
||||
"\n",
|
||||
"Load a classifier that you have trained (specify the classifiers filename) and evaluate it on the satellite images.\n",
|
||||
"\n",
|
||||
"This section will save the output of the classification for each site in a directory named \\evaluation."
|
||||
]
|
||||
},
|
||||
{
|
||||
"cell_type": "code",
|
||||
"execution_count": null,
|
||||
"metadata": {},
|
||||
"outputs": [],
|
||||
"source": [
|
||||
"# load and evaluate a classifier\n",
|
||||
"%matplotlib qt\n",
|
||||
"classifier = joblib.load(os.path.join(filepath_models, 'NN_4classes_Landsat_test.pkl'))\n",
|
||||
"settings['output_epsg'] = 3857\n",
|
||||
"settings['min_beach_area'] = 4500\n",
|
||||
"settings['buffer_size'] = 200\n",
|
||||
"settings['min_length_sl'] = 200\n",
|
||||
"settings['cloud_thresh'] = 0.5\n",
|
||||
"# visualise the classified images\n",
|
||||
"for site in train_sites:\n",
|
||||
" settings['inputs']['sitename'] = site[:site.find('.')] \n",
|
||||
" # load metadata\n",
|
||||
" metadata = SDS_download.get_metadata(settings['inputs'])\n",
|
||||
" # plot the classified images\n",
|
||||
" SDS_classify.evaluate_classifier(classifier,metadata,settings)"
|
||||
]
|
||||
}
|
||||
],
|
||||
"metadata": {
|
||||
"kernelspec": {
|
||||
"display_name": "Python 3",
|
||||
"language": "python",
|
||||
"name": "python3"
|
||||
},
|
||||
"language_info": {
|
||||
"codemirror_mode": {
|
||||
"name": "ipython",
|
||||
"version": 3
|
||||
},
|
||||
"file_extension": ".py",
|
||||
"mimetype": "text/x-python",
|
||||
"name": "python",
|
||||
"nbconvert_exporter": "python",
|
||||
"pygments_lexer": "ipython3",
|
||||
"version": "3.7.3"
|
||||
},
|
||||
"toc": {
|
||||
"base_numbering": 1,
|
||||
"nav_menu": {},
|
||||
"number_sections": false,
|
||||
"sideBar": true,
|
||||
"skip_h1_title": false,
|
||||
"title_cell": "Table of Contents",
|
||||
"title_sidebar": "Contents",
|
||||
"toc_cell": false,
|
||||
"toc_position": {},
|
||||
"toc_section_display": true,
|
||||
"toc_window_display": false
|
||||
},
|
||||
"varInspector": {
|
||||
"cols": {
|
||||
"lenName": 16,
|
||||
"lenType": 16,
|
||||
"lenVar": 40
|
||||
},
|
||||
"kernels_config": {
|
||||
"python": {
|
||||
"delete_cmd_postfix": "",
|
||||
"delete_cmd_prefix": "del ",
|
||||
"library": "var_list.py",
|
||||
"varRefreshCmd": "print(var_dic_list())"
|
||||
},
|
||||
"r": {
|
||||
"delete_cmd_postfix": ") ",
|
||||
"delete_cmd_prefix": "rm(",
|
||||
"library": "var_list.r",
|
||||
"varRefreshCmd": "cat(var_dic_list()) "
|
||||
}
|
||||
},
|
||||
"types_to_exclude": [
|
||||
"module",
|
||||
"function",
|
||||
"builtin_function_or_method",
|
||||
"instance",
|
||||
"_Feature"
|
||||
],
|
||||
"window_display": false
|
||||
}
|
||||
},
|
||||
"nbformat": 4,
|
||||
"nbformat_minor": 2
|
||||
}
|
||||
@ -0,0 +1,36 @@
|
||||
### Train a new CoastSat classifier
|
||||
|
||||
CoastSat's shoreline mapping alogorithm uses an image classification scheme to label each pixel into 4 classes: sand, water, white-water and other land features. While this classifier has been trained using a wide range of different beaches, it may be that it does not perform very well at specific sites that it has never seen before.
|
||||
|
||||
For this reason, we provide the possibility to re-train the classifier by adding labelled data from new sites. This can be done very quickly and easily by using this [Jupyter Notebook](https://github.com/kvos/CoastSat/blob/CoastSat-classifier/classification/train_new_classifier.ipynb).
|
||||
|
||||
Let's take this example, Playa Chañaral in the Atacama desert, Chile. At this beach, the sand is extremely white and the default classifier is not able to label correctly the sand pixels:
|
||||
|
||||
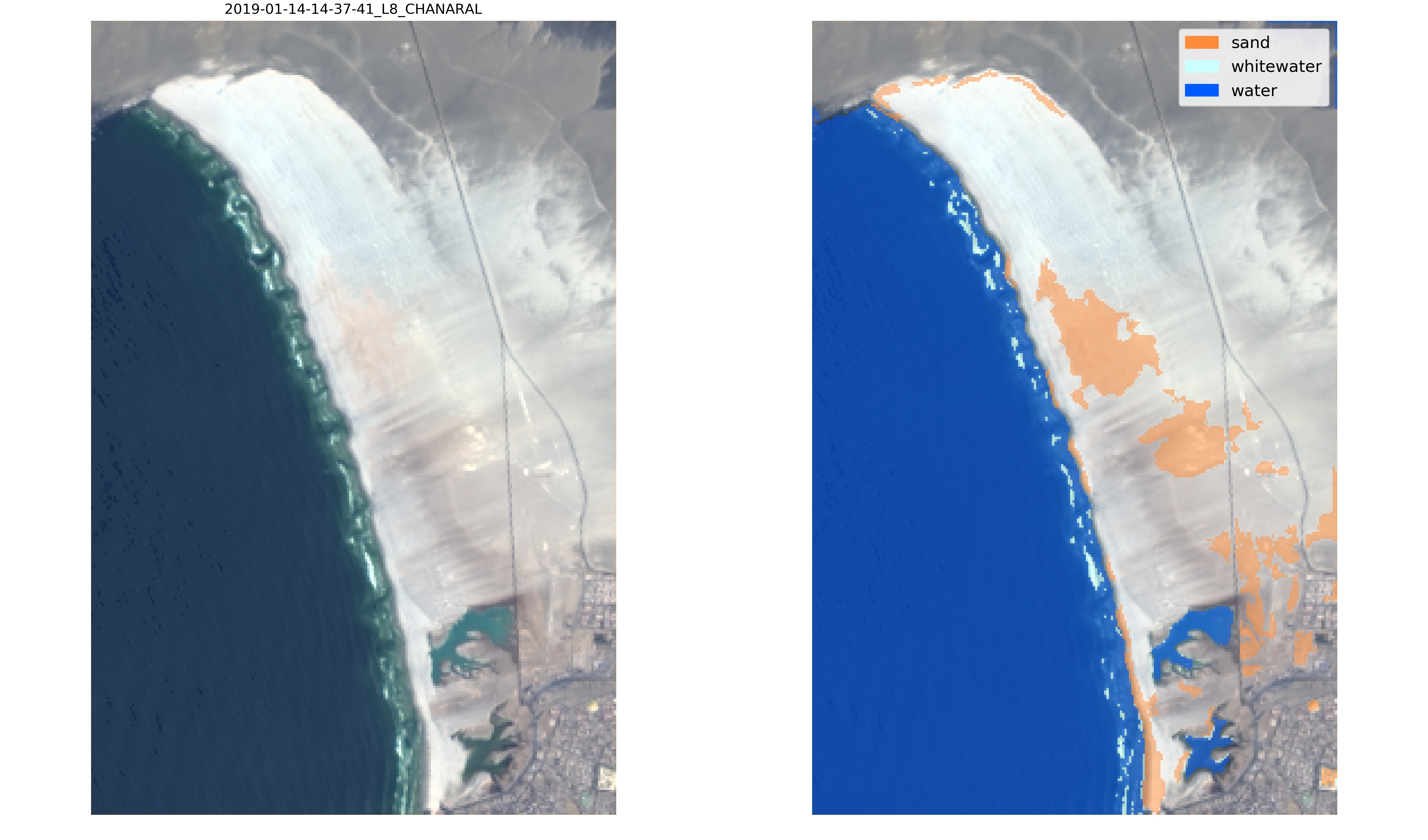
|
||||
|
||||
To overcome this issue, we can generate training data for this site by labelling new images.
|
||||
Download the new images to be labelled and then call the function `SDS_classify.label_images(metadata,settings)`, an interactive tool will pop up for quick and efficient labelling:
|
||||
|
||||

|
||||
|
||||
You only need to label sand pixels, as water and white-water looks the same everywhere in the world. You can label 2-3 images in a few minutes with the interactive tool and then the new labels can be used to re-train the classifier. The labelling tool uses *flood fill* to speed up the selection of sand pixels and you can tune the tolerance of the *flood fill* function in `settings['tolerance']`.
|
||||
|
||||
You can then train a classifier with the newly labelled data.
|
||||
Different classification schemes exist, in this example we use a Multilayer Perceptron (Neural Network) with 2 layers, one of 100 neurons and one of 50 neurons. The training data is first divided in train and split, so that we can evaluate the accuracy of the classifier and plot a confusion matrix.
|
||||
```
|
||||
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, shuffle=True, random_state=0)
|
||||
classifier = MLPClassifier(hidden_layer_sizes=(100,50), solver='adam')
|
||||
classifier.fit(X_train,y_train)
|
||||
print('Accuracy: %0.4f' % classifier.score(X_test,y_test))
|
||||
y_pred = classifier.predict(X_test)
|
||||
label_names = ['other land features','sand','white-water','water']
|
||||
SDS_classify.plot_confusion_matrix(y_test, y_pred,classes=label_names,normalize=False);
|
||||
```
|
||||
|
||||
<img src="https://user-images.githubusercontent.com/7217258/69406723-d9c2eb00-0d56-11ea-9eff-4422dc377638.png" alt="confusion_matrix" width="400"/>
|
||||
|
||||
Finally, the new classifier can be applied to the satellite images, for visual inspection by calling the function `SDS_classify.evaluate_classifier(classifier,metadata,settings)` which will save the classified images in */evaluation*:
|
||||
|
||||
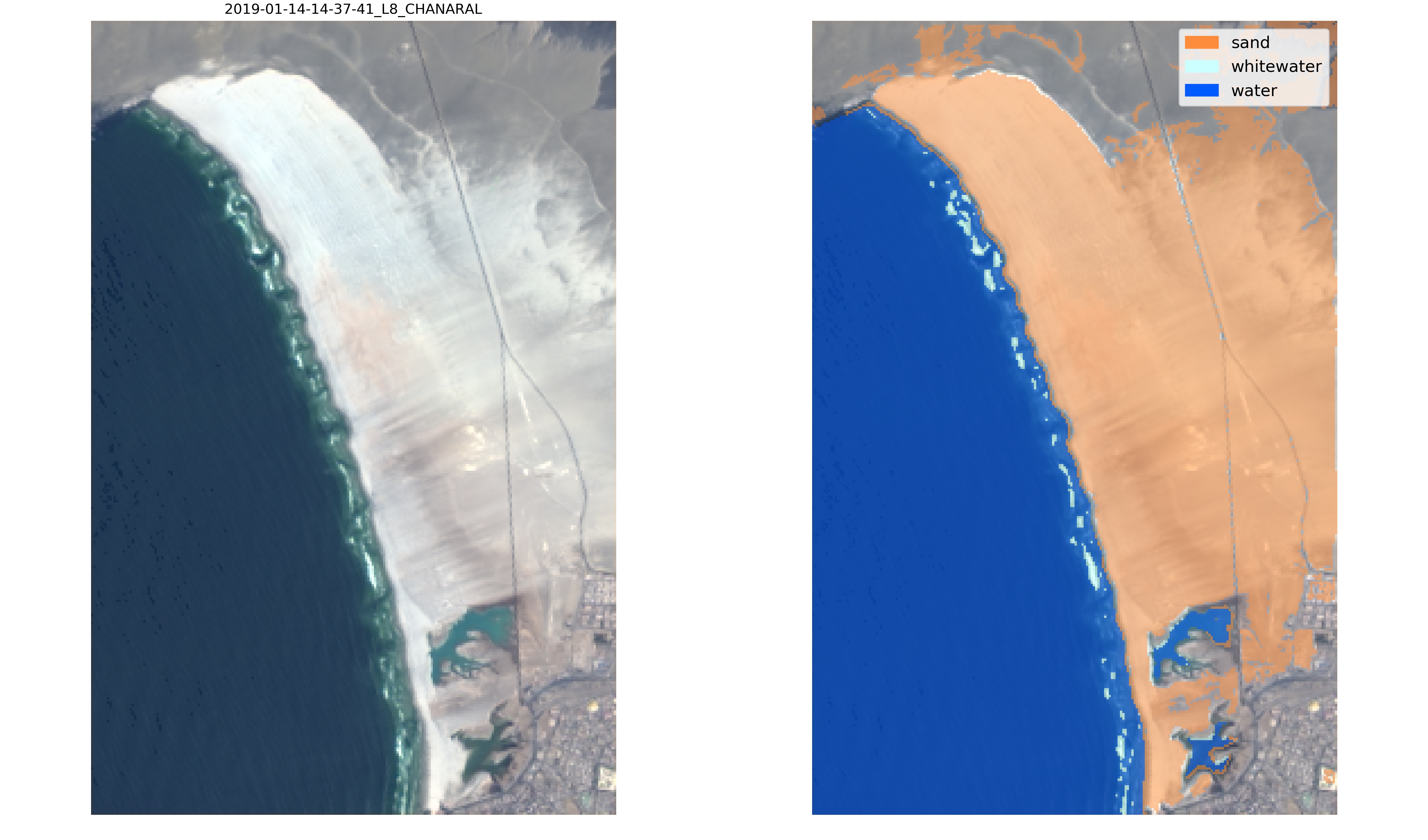
|
||||
|
||||
Now, this new classifier labels correctly the sandy pixels of the Atacama desert and will provide more accurate satellite-derived shorelines at this beach!
|
||||
Binary file not shown.
@ -0,0 +1,62 @@
|
||||
<?xml version="1.0" encoding="UTF-8"?>
|
||||
<kml xmlns="http://www.opengis.net/kml/2.2">
|
||||
<Document>
|
||||
<name>site5</name>
|
||||
<Style id="poly-000000-1200-77-nodesc-normal">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.2</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<Style id="poly-000000-1200-77-nodesc-highlight">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.8</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<StyleMap id="poly-000000-1200-77-nodesc">
|
||||
<Pair>
|
||||
<key>normal</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-normal</styleUrl>
|
||||
</Pair>
|
||||
<Pair>
|
||||
<key>highlight</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-highlight</styleUrl>
|
||||
</Pair>
|
||||
</StyleMap>
|
||||
<Placemark>
|
||||
<name>Polygon</name>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc</styleUrl>
|
||||
<Polygon>
|
||||
<outerBoundaryIs>
|
||||
<LinearRing>
|
||||
<tessellate>1</tessellate>
|
||||
<coordinates>
|
||||
153.6170468,-28.6510018,0
|
||||
153.6134419,-28.6621487,0
|
||||
153.6297498,-28.6665921,0
|
||||
153.6333547,-28.655295,0
|
||||
153.6170468,-28.6510018,0
|
||||
</coordinates>
|
||||
</LinearRing>
|
||||
</outerBoundaryIs>
|
||||
</Polygon>
|
||||
</Placemark>
|
||||
</Document>
|
||||
</kml>
|
||||
@ -0,0 +1,62 @@
|
||||
<?xml version="1.0" encoding="UTF-8"?>
|
||||
<kml xmlns="http://www.opengis.net/kml/2.2">
|
||||
<Document>
|
||||
<name>site2</name>
|
||||
<Style id="poly-000000-1200-77-nodesc-normal">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.2</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<Style id="poly-000000-1200-77-nodesc-highlight">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.8</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<StyleMap id="poly-000000-1200-77-nodesc">
|
||||
<Pair>
|
||||
<key>normal</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-normal</styleUrl>
|
||||
</Pair>
|
||||
<Pair>
|
||||
<key>highlight</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-highlight</styleUrl>
|
||||
</Pair>
|
||||
</StyleMap>
|
||||
<Placemark>
|
||||
<name>Polygon</name>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc</styleUrl>
|
||||
<Polygon>
|
||||
<outerBoundaryIs>
|
||||
<LinearRing>
|
||||
<tessellate>1</tessellate>
|
||||
<coordinates>
|
||||
151.7604354,-32.9330576,0
|
||||
151.7480758,-32.9411254,0
|
||||
151.7612079,-32.953226,0
|
||||
151.7750266,-32.9451592,0
|
||||
151.7604354,-32.9330576,0
|
||||
</coordinates>
|
||||
</LinearRing>
|
||||
</outerBoundaryIs>
|
||||
</Polygon>
|
||||
</Placemark>
|
||||
</Document>
|
||||
</kml>
|
||||
@ -0,0 +1,62 @@
|
||||
<?xml version="1.0" encoding="UTF-8"?>
|
||||
<kml xmlns="http://www.opengis.net/kml/2.2">
|
||||
<Document>
|
||||
<name>site4</name>
|
||||
<Style id="poly-000000-1200-77-nodesc-normal">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.2</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<Style id="poly-000000-1200-77-nodesc-highlight">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.8</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<StyleMap id="poly-000000-1200-77-nodesc">
|
||||
<Pair>
|
||||
<key>normal</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-normal</styleUrl>
|
||||
</Pair>
|
||||
<Pair>
|
||||
<key>highlight</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-highlight</styleUrl>
|
||||
</Pair>
|
||||
</StyleMap>
|
||||
<Placemark>
|
||||
<name>Polygon</name>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc</styleUrl>
|
||||
<Polygon>
|
||||
<outerBoundaryIs>
|
||||
<LinearRing>
|
||||
<tessellate>1</tessellate>
|
||||
<coordinates>
|
||||
153.0949026,-30.3586611,0
|
||||
153.0927568,-30.3715099,0
|
||||
153.1108242,-30.3727688,0
|
||||
153.1124979,-30.3600312,0
|
||||
153.0949026,-30.3586611,0
|
||||
</coordinates>
|
||||
</LinearRing>
|
||||
</outerBoundaryIs>
|
||||
</Polygon>
|
||||
</Placemark>
|
||||
</Document>
|
||||
</kml>
|
||||
@ -0,0 +1,624 @@
|
||||
"""
|
||||
This module contains functions to label satellite images, use the labels to
|
||||
train a pixel-wise classifier and evaluate the classifier
|
||||
|
||||
Author: Kilian Vos, Water Research Laboratory, University of New South Wales
|
||||
"""
|
||||
|
||||
# load modules
|
||||
import os
|
||||
import numpy as np
|
||||
import matplotlib.pyplot as plt
|
||||
import matplotlib.cm as cm
|
||||
from matplotlib.widgets import LassoSelector
|
||||
from matplotlib import path
|
||||
import pickle
|
||||
import pdb
|
||||
import warnings
|
||||
warnings.filterwarnings("ignore")
|
||||
|
||||
# image processing modules
|
||||
from skimage.segmentation import flood
|
||||
from skimage import morphology
|
||||
from pylab import ginput
|
||||
from sklearn.metrics import confusion_matrix
|
||||
np.set_printoptions(precision=2)
|
||||
|
||||
# CoastSat modules
|
||||
from coastsat import SDS_preprocess, SDS_shoreline, SDS_tools
|
||||
|
||||
class SelectFromImage(object):
|
||||
"""
|
||||
Class used to draw the lassos on the images with two methods:
|
||||
- onselect: save the pixels inside the selection
|
||||
- disconnect: stop drawing lassos on the image
|
||||
"""
|
||||
# initialize lasso selection class
|
||||
def __init__(self, ax, implot, color=[1,1,1]):
|
||||
self.canvas = ax.figure.canvas
|
||||
self.implot = implot
|
||||
self.array = implot.get_array()
|
||||
xv, yv = np.meshgrid(np.arange(self.array.shape[1]),np.arange(self.array.shape[0]))
|
||||
self.pix = np.vstack( (xv.flatten(), yv.flatten()) ).T
|
||||
self.ind = []
|
||||
self.im_bool = np.zeros((self.array.shape[0], self.array.shape[1]))
|
||||
self.color = color
|
||||
self.lasso = LassoSelector(ax, onselect=self.onselect)
|
||||
|
||||
def onselect(self, verts):
|
||||
# find pixels contained in the lasso
|
||||
p = path.Path(verts)
|
||||
self.ind = p.contains_points(self.pix, radius=1)
|
||||
# color selected pixels
|
||||
array_list = []
|
||||
for k in range(self.array.shape[2]):
|
||||
array2d = self.array[:,:,k]
|
||||
lin = np.arange(array2d.size)
|
||||
new_array2d = array2d.flatten()
|
||||
new_array2d[lin[self.ind]] = self.color[k]
|
||||
array_list.append(new_array2d.reshape(array2d.shape))
|
||||
self.array = np.stack(array_list,axis=2)
|
||||
self.implot.set_data(self.array)
|
||||
self.canvas.draw_idle()
|
||||
# update boolean image with selected pixels
|
||||
vec_bool = self.im_bool.flatten()
|
||||
vec_bool[lin[self.ind]] = 1
|
||||
self.im_bool = vec_bool.reshape(self.im_bool.shape)
|
||||
|
||||
def disconnect(self):
|
||||
self.lasso.disconnect_events()
|
||||
|
||||
def label_images(metadata,settings):
|
||||
"""
|
||||
Load satellite images and interactively label different classes (hard-coded)
|
||||
|
||||
KV WRL 2019
|
||||
|
||||
Arguments:
|
||||
-----------
|
||||
metadata: dict
|
||||
contains all the information about the satellite images that were downloaded
|
||||
settings: dict with the following keys
|
||||
'cloud_thresh': float
|
||||
value between 0 and 1 indicating the maximum cloud fraction in
|
||||
the cropped image that is accepted
|
||||
'cloud_mask_issue': boolean
|
||||
True if there is an issue with the cloud mask and sand pixels
|
||||
are erroneously being masked on the images
|
||||
'labels': dict
|
||||
list of label names (key) and label numbers (value) for each class
|
||||
'flood_fill': boolean
|
||||
True to use the flood_fill functionality when labelling sand pixels
|
||||
'tolerance': float
|
||||
tolerance value for flood fill when labelling the sand pixels
|
||||
'filepath_train': str
|
||||
directory in which to save the labelled data
|
||||
'inputs': dict
|
||||
input parameters (sitename, filepath, polygon, dates, sat_list)
|
||||
|
||||
Returns:
|
||||
-----------
|
||||
Stores the labelled data in the specified directory
|
||||
|
||||
"""
|
||||
|
||||
filepath_train = settings['filepath_train']
|
||||
# initialize figure
|
||||
fig,ax = plt.subplots(1,1,figsize=[17,10], tight_layout=True,sharex=True,
|
||||
sharey=True)
|
||||
mng = plt.get_current_fig_manager()
|
||||
mng.window.showMaximized()
|
||||
|
||||
# loop through satellites
|
||||
for satname in metadata.keys():
|
||||
filepath = SDS_tools.get_filepath(settings['inputs'],satname)
|
||||
filenames = metadata[satname]['filenames']
|
||||
# loop through images
|
||||
for i in range(len(filenames)):
|
||||
# image filename
|
||||
fn = SDS_tools.get_filenames(filenames[i],filepath, satname)
|
||||
# read and preprocess image
|
||||
im_ms, georef, cloud_mask, im_extra, im_QA, im_nodata = SDS_preprocess.preprocess_single(fn, satname, settings['cloud_mask_issue'])
|
||||
# calculate cloud cover
|
||||
cloud_cover = np.divide(sum(sum(cloud_mask.astype(int))),
|
||||
(cloud_mask.shape[0]*cloud_mask.shape[1]))
|
||||
# skip image if cloud cover is above threshold
|
||||
if cloud_cover > settings['cloud_thresh'] or cloud_cover == 1:
|
||||
continue
|
||||
# get individual RGB image
|
||||
im_RGB = SDS_preprocess.rescale_image_intensity(im_ms[:,:,[2,1,0]], cloud_mask, 99.9)
|
||||
im_NDVI = SDS_tools.nd_index(im_ms[:,:,3], im_ms[:,:,2], cloud_mask)
|
||||
im_NDWI = SDS_tools.nd_index(im_ms[:,:,3], im_ms[:,:,1], cloud_mask)
|
||||
# initialise labels
|
||||
im_viz = im_RGB.copy()
|
||||
im_labels = np.zeros([im_RGB.shape[0],im_RGB.shape[1]])
|
||||
# show RGB image
|
||||
ax.axis('off')
|
||||
ax.imshow(im_RGB)
|
||||
implot = ax.imshow(im_viz, alpha=0.6)
|
||||
filename = filenames[i][:filenames[i].find('.')][:-4]
|
||||
ax.set_title(filename)
|
||||
|
||||
##############################################################
|
||||
# select image to label
|
||||
##############################################################
|
||||
# set a key event to accept/reject the detections (see https://stackoverflow.com/a/15033071)
|
||||
# this variable needs to be immuatable so we can access it after the keypress event
|
||||
key_event = {}
|
||||
def press(event):
|
||||
# store what key was pressed in the dictionary
|
||||
key_event['pressed'] = event.key
|
||||
# let the user press a key, right arrow to keep the image, left arrow to skip it
|
||||
# to break the loop the user can press 'escape'
|
||||
while True:
|
||||
btn_keep = ax.text(1.1, 0.9, 'keep ⇨', size=12, ha="right", va="top",
|
||||
transform=ax.transAxes,
|
||||
bbox=dict(boxstyle="square", ec='k',fc='w'))
|
||||
btn_skip = ax.text(-0.1, 0.9, '⇦ skip', size=12, ha="left", va="top",
|
||||
transform=ax.transAxes,
|
||||
bbox=dict(boxstyle="square", ec='k',fc='w'))
|
||||
btn_esc = ax.text(0.5, 0, '<esc> to quit', size=12, ha="center", va="top",
|
||||
transform=ax.transAxes,
|
||||
bbox=dict(boxstyle="square", ec='k',fc='w'))
|
||||
fig.canvas.draw_idle()
|
||||
fig.canvas.mpl_connect('key_press_event', press)
|
||||
plt.waitforbuttonpress()
|
||||
# after button is pressed, remove the buttons
|
||||
btn_skip.remove()
|
||||
btn_keep.remove()
|
||||
btn_esc.remove()
|
||||
|
||||
# keep/skip image according to the pressed key, 'escape' to break the loop
|
||||
if key_event.get('pressed') == 'right':
|
||||
skip_image = False
|
||||
break
|
||||
elif key_event.get('pressed') == 'left':
|
||||
skip_image = True
|
||||
break
|
||||
elif key_event.get('pressed') == 'escape':
|
||||
plt.close()
|
||||
raise StopIteration('User cancelled labelling images')
|
||||
else:
|
||||
plt.waitforbuttonpress()
|
||||
|
||||
# if user decided to skip show the next image
|
||||
if skip_image:
|
||||
ax.clear()
|
||||
continue
|
||||
# otherwise label this image
|
||||
else:
|
||||
##############################################################
|
||||
# digitize sandy pixels
|
||||
##############################################################
|
||||
ax.set_title('Click on SAND pixels (flood fill activated, tolerance = %.2f)\nwhen finished press <Enter>'%settings['tolerance'])
|
||||
# create erase button, if you click there it delets the last selection
|
||||
btn_erase = ax.text(im_ms.shape[1], 0, 'Erase', size=20, ha='right', va='top',
|
||||
bbox=dict(boxstyle="square", ec='k',fc='w'))
|
||||
fig.canvas.draw_idle()
|
||||
color_sand = settings['colors']['sand']
|
||||
sand_pixels = []
|
||||
while 1:
|
||||
seed = ginput(n=1, timeout=0, show_clicks=True)
|
||||
# if empty break the loop and go to next label
|
||||
if len(seed) == 0:
|
||||
break
|
||||
else:
|
||||
# round to pixel location
|
||||
seed = np.round(seed[0]).astype(int)
|
||||
# if user clicks on erase, delete the last selection
|
||||
if seed[0] > 0.95*im_ms.shape[1] and seed[1] < 0.05*im_ms.shape[0]:
|
||||
if len(sand_pixels) > 0:
|
||||
im_labels[sand_pixels[-1]] = 0
|
||||
for k in range(im_viz.shape[2]):
|
||||
im_viz[sand_pixels[-1],k] = im_RGB[sand_pixels[-1],k]
|
||||
implot.set_data(im_viz)
|
||||
fig.canvas.draw_idle()
|
||||
del sand_pixels[-1]
|
||||
|
||||
# otherwise label the selected sand pixels
|
||||
else:
|
||||
# flood fill the NDVI and the NDWI
|
||||
fill_NDVI = flood(im_NDVI, (seed[1],seed[0]), tolerance=settings['tolerance'])
|
||||
fill_NDWI = flood(im_NDWI, (seed[1],seed[0]), tolerance=settings['tolerance'])
|
||||
# compute the intersection of the two masks
|
||||
fill_sand = np.logical_and(fill_NDVI, fill_NDWI)
|
||||
im_labels[fill_sand] = settings['labels']['sand']
|
||||
sand_pixels.append(fill_sand)
|
||||
# show the labelled pixels
|
||||
for k in range(im_viz.shape[2]):
|
||||
im_viz[im_labels==settings['labels']['sand'],k] = color_sand[k]
|
||||
implot.set_data(im_viz)
|
||||
fig.canvas.draw_idle()
|
||||
|
||||
##############################################################
|
||||
# digitize white-water pixels
|
||||
##############################################################
|
||||
color_ww = settings['colors']['white-water']
|
||||
ax.set_title('Click on individual WHITE-WATER pixels (no flood fill)\nwhen finished press <Enter>')
|
||||
fig.canvas.draw_idle()
|
||||
ww_pixels = []
|
||||
while 1:
|
||||
seed = ginput(n=1, timeout=0, show_clicks=True)
|
||||
# if empty break the loop and go to next label
|
||||
if len(seed) == 0:
|
||||
break
|
||||
else:
|
||||
# round to pixel location
|
||||
seed = np.round(seed[0]).astype(int)
|
||||
# if user clicks on erase, delete the last labelled pixels
|
||||
if seed[0] > 0.95*im_ms.shape[1] and seed[1] < 0.05*im_ms.shape[0]:
|
||||
if len(ww_pixels) > 0:
|
||||
im_labels[ww_pixels[-1][1],ww_pixels[-1][0]] = 0
|
||||
for k in range(im_viz.shape[2]):
|
||||
im_viz[ww_pixels[-1][1],ww_pixels[-1][0],k] = im_RGB[ww_pixels[-1][1],ww_pixels[-1][0],k]
|
||||
implot.set_data(im_viz)
|
||||
fig.canvas.draw_idle()
|
||||
del ww_pixels[-1]
|
||||
else:
|
||||
im_labels[seed[1],seed[0]] = settings['labels']['white-water']
|
||||
for k in range(im_viz.shape[2]):
|
||||
im_viz[seed[1],seed[0],k] = color_ww[k]
|
||||
implot.set_data(im_viz)
|
||||
fig.canvas.draw_idle()
|
||||
ww_pixels.append(seed)
|
||||
|
||||
im_sand_ww = im_viz.copy()
|
||||
btn_erase.set(text='<Esc> to Erase', fontsize=12)
|
||||
|
||||
##############################################################
|
||||
# digitize water pixels (with lassos)
|
||||
##############################################################
|
||||
color_water = settings['colors']['water']
|
||||
ax.set_title('Click and hold to draw lassos and select WATER pixels\nwhen finished press <Enter>')
|
||||
fig.canvas.draw_idle()
|
||||
selector_water = SelectFromImage(ax, implot, color_water)
|
||||
key_event = {}
|
||||
while True:
|
||||
fig.canvas.draw_idle()
|
||||
fig.canvas.mpl_connect('key_press_event', press)
|
||||
plt.waitforbuttonpress()
|
||||
if key_event.get('pressed') == 'enter':
|
||||
selector_water.disconnect()
|
||||
break
|
||||
elif key_event.get('pressed') == 'escape':
|
||||
selector_water.array = im_sand_ww
|
||||
implot.set_data(selector_water.array)
|
||||
fig.canvas.draw_idle()
|
||||
selector_water.implot = implot
|
||||
selector_water.im_bool = np.zeros((selector_water.array.shape[0], selector_water.array.shape[1]))
|
||||
selector_water.ind=[]
|
||||
# update im_viz and im_labels
|
||||
im_viz = selector_water.array
|
||||
selector_water.im_bool = selector_water.im_bool.astype(bool)
|
||||
im_labels[selector_water.im_bool] = settings['labels']['water']
|
||||
|
||||
im_sand_ww_water = im_viz.copy()
|
||||
|
||||
##############################################################
|
||||
# digitize land pixels (with lassos)
|
||||
##############################################################
|
||||
color_land = settings['colors']['other land features']
|
||||
ax.set_title('Click and hold to draw lassos and select OTHER LAND pixels\nwhen finished press <Enter>')
|
||||
fig.canvas.draw_idle()
|
||||
selector_land = SelectFromImage(ax, implot, color_land)
|
||||
key_event = {}
|
||||
while True:
|
||||
fig.canvas.draw_idle()
|
||||
fig.canvas.mpl_connect('key_press_event', press)
|
||||
plt.waitforbuttonpress()
|
||||
if key_event.get('pressed') == 'enter':
|
||||
selector_land.disconnect()
|
||||
break
|
||||
elif key_event.get('pressed') == 'escape':
|
||||
selector_land.array = im_sand_ww_water
|
||||
implot.set_data(selector_land.array)
|
||||
fig.canvas.draw_idle()
|
||||
selector_land.implot = implot
|
||||
selector_land.im_bool = np.zeros((selector_land.array.shape[0], selector_land.array.shape[1]))
|
||||
selector_land.ind=[]
|
||||
# update im_viz and im_labels
|
||||
im_viz = selector_land.array
|
||||
selector_land.im_bool = selector_land.im_bool.astype(bool)
|
||||
im_labels[selector_land.im_bool] = settings['labels']['other land features']
|
||||
|
||||
# save labelled image
|
||||
ax.set_title(filename)
|
||||
fig.canvas.draw_idle()
|
||||
fp = os.path.join(filepath_train,settings['inputs']['sitename'])
|
||||
if not os.path.exists(fp):
|
||||
os.makedirs(fp)
|
||||
fig.savefig(os.path.join(fp,filename+'.jpg'), dpi=150)
|
||||
ax.clear()
|
||||
# save labels and features
|
||||
features = dict([])
|
||||
for key in settings['labels'].keys():
|
||||
im_bool = im_labels == settings['labels'][key]
|
||||
features[key] = SDS_shoreline.calculate_features(im_ms, cloud_mask, im_bool)
|
||||
training_data = {'labels':im_labels, 'features':features, 'label_ids':settings['labels']}
|
||||
with open(os.path.join(fp, filename + '.pkl'), 'wb') as f:
|
||||
pickle.dump(training_data,f)
|
||||
|
||||
# close figure when finished
|
||||
plt.close(fig)
|
||||
|
||||
def load_labels(train_sites, settings):
|
||||
"""
|
||||
Load the labelled data from the different training sites
|
||||
|
||||
KV WRL 2019
|
||||
|
||||
Arguments:
|
||||
-----------
|
||||
train_sites: list of str
|
||||
sites to be loaded
|
||||
settings: dict with the following keys
|
||||
'labels': dict
|
||||
list of label names (key) and label numbers (value) for each class
|
||||
'filepath_train': str
|
||||
directory in which to save the labelled data
|
||||
|
||||
Returns:
|
||||
-----------
|
||||
features: dict
|
||||
contains the features for each labelled pixel
|
||||
|
||||
"""
|
||||
|
||||
filepath_train = settings['filepath_train']
|
||||
# initialize the features dict
|
||||
features = dict([])
|
||||
n_features = 20
|
||||
first_row = np.nan*np.ones((1,n_features))
|
||||
for key in settings['labels'].keys():
|
||||
features[key] = first_row
|
||||
# loop through each site
|
||||
for site in train_sites:
|
||||
sitename = site[:site.find('.')]
|
||||
filepath = os.path.join(filepath_train,sitename)
|
||||
if os.path.exists(filepath):
|
||||
list_files = os.listdir(filepath)
|
||||
else:
|
||||
continue
|
||||
# make a new list with only the .pkl files (no .jpg)
|
||||
list_files_pkl = []
|
||||
for file in list_files:
|
||||
if '.pkl' in file:
|
||||
list_files_pkl.append(file)
|
||||
# load and append the training data to the features dict
|
||||
for file in list_files_pkl:
|
||||
# read file
|
||||
with open(os.path.join(filepath, file), 'rb') as f:
|
||||
labelled_data = pickle.load(f)
|
||||
for key in labelled_data['features'].keys():
|
||||
if len(labelled_data['features'][key])>0: # check that is not empty
|
||||
# append rows
|
||||
features[key] = np.append(features[key],
|
||||
labelled_data['features'][key], axis=0)
|
||||
# remove the first row (initialized with nans) and print how many pixels
|
||||
print('Number of pixels per class in training data:')
|
||||
for key in features.keys():
|
||||
features[key] = features[key][1:,:]
|
||||
print('%s : %d pixels'%(key,len(features[key])))
|
||||
|
||||
return features
|
||||
|
||||
def format_training_data(features, classes, labels):
|
||||
"""
|
||||
Format the labelled data in an X features matrix and a y labels vector, so
|
||||
that it can be used for training an ML model.
|
||||
|
||||
KV WRL 2019
|
||||
|
||||
Arguments:
|
||||
-----------
|
||||
features: dict
|
||||
contains the features for each labelled pixel
|
||||
classes: list of str
|
||||
names of the classes
|
||||
labels: list of int
|
||||
int value associated with each class (in the same order as classes)
|
||||
|
||||
Returns:
|
||||
-----------
|
||||
X: np.array
|
||||
matrix features along the columns and pixels along the rows
|
||||
y: np.array
|
||||
vector with the labels corresponding to each row of X
|
||||
|
||||
"""
|
||||
|
||||
# initialize X and y
|
||||
X = np.nan*np.ones((1,features[classes[0]].shape[1]))
|
||||
y = np.nan*np.ones((1,1))
|
||||
# append row of features to X and corresponding label to y
|
||||
for i,key in enumerate(classes):
|
||||
y = np.append(y, labels[i]*np.ones((features[key].shape[0],1)), axis=0)
|
||||
X = np.append(X, features[key], axis=0)
|
||||
# remove first row
|
||||
X = X[1:,:]; y = y[1:]
|
||||
# replace nans with something close to 0
|
||||
# training algotihms cannot handle nans
|
||||
X[np.isnan(X)] = 1e-9
|
||||
|
||||
return X, y
|
||||
|
||||
def plot_confusion_matrix(y_true,y_pred,classes,normalize=False,cmap=plt.cm.Blues):
|
||||
"""
|
||||
Function copied from the scikit-learn examples (https://scikit-learn.org/stable/)
|
||||
This function plots a confusion matrix.
|
||||
Normalization can be applied by setting `normalize=True`.
|
||||
|
||||
"""
|
||||
# compute confusion matrix
|
||||
cm = confusion_matrix(y_true, y_pred)
|
||||
if normalize:
|
||||
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
|
||||
print("Normalized confusion matrix")
|
||||
else:
|
||||
print('Confusion matrix, without normalization')
|
||||
|
||||
# plot confusion matrix
|
||||
fig, ax = plt.subplots(figsize=(6,6), tight_layout=True)
|
||||
im = ax.imshow(cm, interpolation='nearest', cmap=cmap)
|
||||
# ax.figure.colorbar(im, ax=ax)
|
||||
ax.set(xticks=np.arange(cm.shape[1]),
|
||||
yticks=np.arange(cm.shape[0]), ylim=[3.5,-0.5],
|
||||
xticklabels=classes, yticklabels=classes,
|
||||
ylabel='True label',
|
||||
xlabel='Predicted label')
|
||||
|
||||
# rotate the tick labels and set their alignment.
|
||||
plt.setp(ax.get_xticklabels(), rotation=45, ha="right",
|
||||
rotation_mode="anchor")
|
||||
|
||||
# loop over data dimensions and create text annotations.
|
||||
fmt = '.2f' if normalize else 'd'
|
||||
thresh = cm.max() / 2.
|
||||
for i in range(cm.shape[0]):
|
||||
for j in range(cm.shape[1]):
|
||||
ax.text(j, i, format(cm[i, j], fmt),
|
||||
ha="center", va="center",
|
||||
color="white" if cm[i, j] > thresh else "black",
|
||||
fontsize=12)
|
||||
fig.tight_layout()
|
||||
return ax
|
||||
|
||||
def evaluate_classifier(classifier, metadata, settings):
|
||||
"""
|
||||
Apply the image classifier to all the images and save the classified images.
|
||||
|
||||
KV WRL 2019
|
||||
|
||||
Arguments:
|
||||
-----------
|
||||
classifier: joblib object
|
||||
classifier model to be used for image classification
|
||||
metadata: dict
|
||||
contains all the information about the satellite images that were downloaded
|
||||
settings: dict with the following keys
|
||||
'inputs': dict
|
||||
input parameters (sitename, filepath, polygon, dates, sat_list)
|
||||
'cloud_thresh': float
|
||||
value between 0 and 1 indicating the maximum cloud fraction in
|
||||
the cropped image that is accepted
|
||||
'cloud_mask_issue': boolean
|
||||
True if there is an issue with the cloud mask and sand pixels
|
||||
are erroneously being masked on the images
|
||||
'output_epsg': int
|
||||
output spatial reference system as EPSG code
|
||||
'buffer_size': int
|
||||
size of the buffer (m) around the sandy pixels over which the pixels
|
||||
are considered in the thresholding algorithm
|
||||
'min_beach_area': int
|
||||
minimum allowable object area (in metres^2) for the class 'sand',
|
||||
the area is converted to number of connected pixels
|
||||
'min_length_sl': int
|
||||
minimum length (in metres) of shoreline contour to be valid
|
||||
|
||||
Returns:
|
||||
-----------
|
||||
Saves .jpg images with the output of the classification in the folder ./detection
|
||||
|
||||
"""
|
||||
|
||||
# create folder called evaluation
|
||||
fp = os.path.join(os.getcwd(), 'evaluation')
|
||||
if not os.path.exists(fp):
|
||||
os.makedirs(fp)
|
||||
|
||||
# initialize figure (not interactive)
|
||||
plt.ioff()
|
||||
fig,ax = plt.subplots(1,2,figsize=[17,10],sharex=True, sharey=True,
|
||||
constrained_layout=True)
|
||||
|
||||
# create colormap for labels
|
||||
cmap = cm.get_cmap('tab20c')
|
||||
colorpalette = cmap(np.arange(0,13,1))
|
||||
colours = np.zeros((3,4))
|
||||
colours[0,:] = colorpalette[5]
|
||||
colours[1,:] = np.array([204/255,1,1,1])
|
||||
colours[2,:] = np.array([0,91/255,1,1])
|
||||
# loop through satellites
|
||||
for satname in metadata.keys():
|
||||
filepath = SDS_tools.get_filepath(settings['inputs'],satname)
|
||||
filenames = metadata[satname]['filenames']
|
||||
|
||||
# load classifiers and
|
||||
if satname in ['L5','L7','L8']:
|
||||
pixel_size = 15
|
||||
elif satname == 'S2':
|
||||
pixel_size = 10
|
||||
# convert settings['min_beach_area'] and settings['buffer_size'] from metres to pixels
|
||||
buffer_size_pixels = np.ceil(settings['buffer_size']/pixel_size)
|
||||
min_beach_area_pixels = np.ceil(settings['min_beach_area']/pixel_size**2)
|
||||
|
||||
# loop through images
|
||||
for i in range(len(filenames)):
|
||||
# image filename
|
||||
fn = SDS_tools.get_filenames(filenames[i],filepath, satname)
|
||||
# read and preprocess image
|
||||
im_ms, georef, cloud_mask, im_extra, im_QA, im_nodata = SDS_preprocess.preprocess_single(fn, satname, settings['cloud_mask_issue'])
|
||||
image_epsg = metadata[satname]['epsg'][i]
|
||||
# calculate cloud cover
|
||||
cloud_cover = np.divide(sum(sum(cloud_mask.astype(int))),
|
||||
(cloud_mask.shape[0]*cloud_mask.shape[1]))
|
||||
# skip image if cloud cover is above threshold
|
||||
if cloud_cover > settings['cloud_thresh']:
|
||||
continue
|
||||
# calculate a buffer around the reference shoreline (if any has been digitised)
|
||||
im_ref_buffer = SDS_shoreline.create_shoreline_buffer(cloud_mask.shape, georef, image_epsg,
|
||||
pixel_size, settings)
|
||||
# classify image in 4 classes (sand, whitewater, water, other) with NN classifier
|
||||
im_classif, im_labels = SDS_shoreline.classify_image_NN(im_ms, im_extra, cloud_mask,
|
||||
min_beach_area_pixels, classifier)
|
||||
# there are two options to map the contours:
|
||||
# if there are pixels in the 'sand' class --> use find_wl_contours2 (enhanced)
|
||||
# otherwise use find_wl_contours2 (traditional)
|
||||
try: # use try/except structure for long runs
|
||||
if sum(sum(im_labels[:,:,0])) < 10 :
|
||||
# compute MNDWI image (SWIR-G)
|
||||
im_mndwi = SDS_tools.nd_index(im_ms[:,:,4], im_ms[:,:,1], cloud_mask)
|
||||
# find water contours on MNDWI grayscale image
|
||||
contours_mwi = SDS_shoreline.find_wl_contours1(im_mndwi, cloud_mask, im_ref_buffer)
|
||||
else:
|
||||
# use classification to refine threshold and extract the sand/water interface
|
||||
contours_wi, contours_mwi = SDS_shoreline.find_wl_contours2(im_ms, im_labels,
|
||||
cloud_mask, buffer_size_pixels, im_ref_buffer)
|
||||
except:
|
||||
print('Could not map shoreline for this image: ' + filenames[i])
|
||||
continue
|
||||
# process the water contours into a shoreline
|
||||
shoreline = SDS_shoreline.process_shoreline(contours_mwi, cloud_mask, georef, image_epsg, settings)
|
||||
try:
|
||||
sl_pix = SDS_tools.convert_world2pix(SDS_tools.convert_epsg(shoreline,
|
||||
settings['output_epsg'],
|
||||
image_epsg)[:,[0,1]], georef)
|
||||
except:
|
||||
# if try fails, just add nan into the shoreline vector so the next parts can still run
|
||||
sl_pix = np.array([[np.nan, np.nan],[np.nan, np.nan]])
|
||||
# make a plot
|
||||
im_RGB = SDS_preprocess.rescale_image_intensity(im_ms[:,:,[2,1,0]], cloud_mask, 99.9)
|
||||
# create classified image
|
||||
im_class = np.copy(im_RGB)
|
||||
for k in range(0,im_labels.shape[2]):
|
||||
im_class[im_labels[:,:,k],0] = colours[k,0]
|
||||
im_class[im_labels[:,:,k],1] = colours[k,1]
|
||||
im_class[im_labels[:,:,k],2] = colours[k,2]
|
||||
# show images
|
||||
ax[0].imshow(im_RGB)
|
||||
ax[1].imshow(im_RGB)
|
||||
ax[1].imshow(im_class, alpha=0.5)
|
||||
ax[0].axis('off')
|
||||
ax[1].axis('off')
|
||||
filename = filenames[i][:filenames[i].find('.')][:-4]
|
||||
ax[0].set_title(filename)
|
||||
ax[0].plot(sl_pix[:,0], sl_pix[:,1], 'k.', markersize=3)
|
||||
ax[1].plot(sl_pix[:,0], sl_pix[:,1], 'k.', markersize=3)
|
||||
# save figure
|
||||
fig.savefig(os.path.join(fp,settings['inputs']['sitename'] + filename[:19] +'.jpg'), dpi=150)
|
||||
# clear axes
|
||||
for cax in fig.axes:
|
||||
cax.clear()
|
||||
|
||||
# close the figure at the end
|
||||
plt.close()
|
||||
@ -0,0 +1,62 @@
|
||||
<?xml version="1.0" encoding="UTF-8"?>
|
||||
<kml xmlns="http://www.opengis.net/kml/2.2">
|
||||
<Document>
|
||||
<name>NARRA</name>
|
||||
<Style id="poly-000000-1200-77-nodesc-normal">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.2</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<Style id="poly-000000-1200-77-nodesc-highlight">
|
||||
<LineStyle>
|
||||
<color>ff000000</color>
|
||||
<width>1.8</width>
|
||||
</LineStyle>
|
||||
<PolyStyle>
|
||||
<color>4d000000</color>
|
||||
<fill>1</fill>
|
||||
<outline>1</outline>
|
||||
</PolyStyle>
|
||||
<BalloonStyle>
|
||||
<text><![CDATA[<h3>$[name]</h3>]]></text>
|
||||
</BalloonStyle>
|
||||
</Style>
|
||||
<StyleMap id="poly-000000-1200-77-nodesc">
|
||||
<Pair>
|
||||
<key>normal</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-normal</styleUrl>
|
||||
</Pair>
|
||||
<Pair>
|
||||
<key>highlight</key>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc-highlight</styleUrl>
|
||||
</Pair>
|
||||
</StyleMap>
|
||||
<Placemark>
|
||||
<name>Polygon 1</name>
|
||||
<styleUrl>#poly-000000-1200-77-nodesc</styleUrl>
|
||||
<Polygon>
|
||||
<outerBoundaryIs>
|
||||
<LinearRing>
|
||||
<tessellate>1</tessellate>
|
||||
<coordinates>
|
||||
151.2957545,-33.7012561,0
|
||||
151.297557,-33.7388075,0
|
||||
151.312234,-33.7390216,0
|
||||
151.311204,-33.701399,0
|
||||
151.2957545,-33.7012561,0
|
||||
</coordinates>
|
||||
</LinearRing>
|
||||
</outerBoundaryIs>
|
||||
</Polygon>
|
||||
</Placemark>
|
||||
</Document>
|
||||
</kml>
|
||||
Loading…
Reference in New Issue